我們今天玩的是AutoML Natural Language,可以透過GCP左邊的sidebar,找到Natural Language打開,或直接點這個連結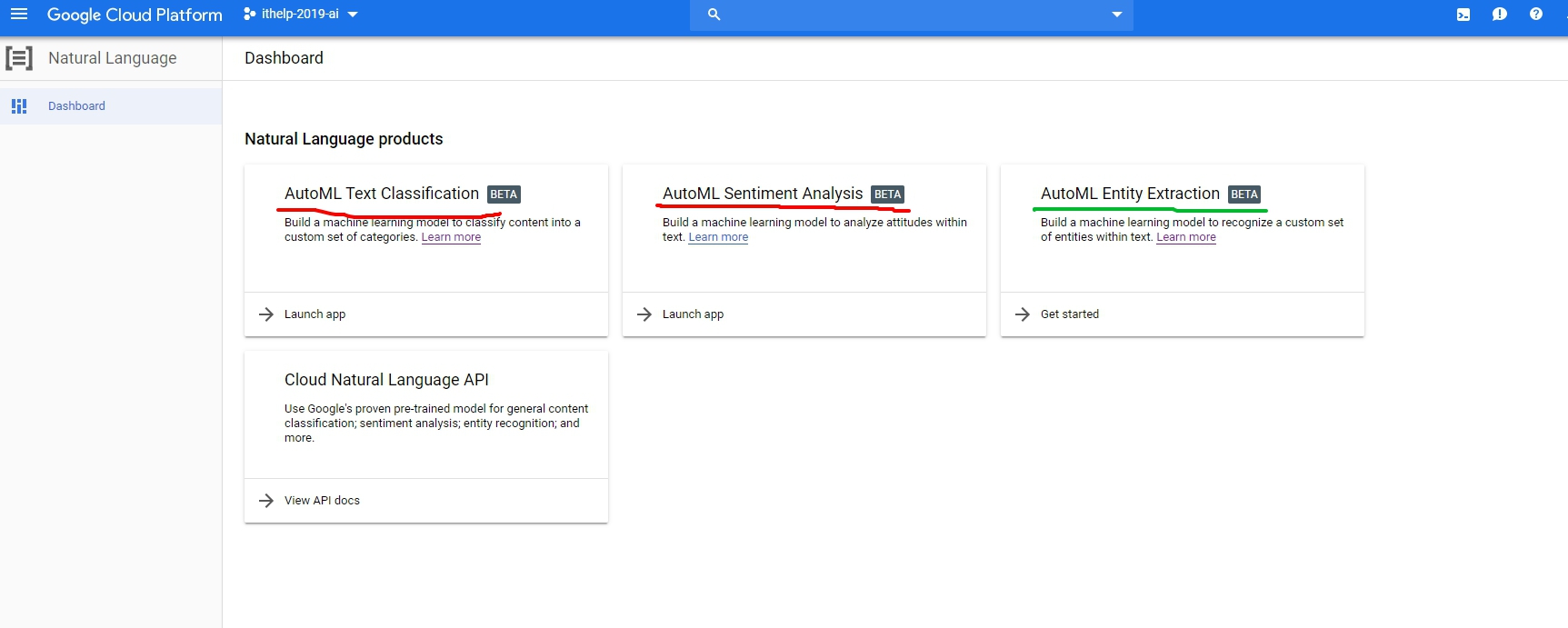
AutoML Natural Language的收費方式記得先看這裡:https://cloud.google.com/natural-language/automl/pricing?_ga=2.125750730.-1764044900.1562297905
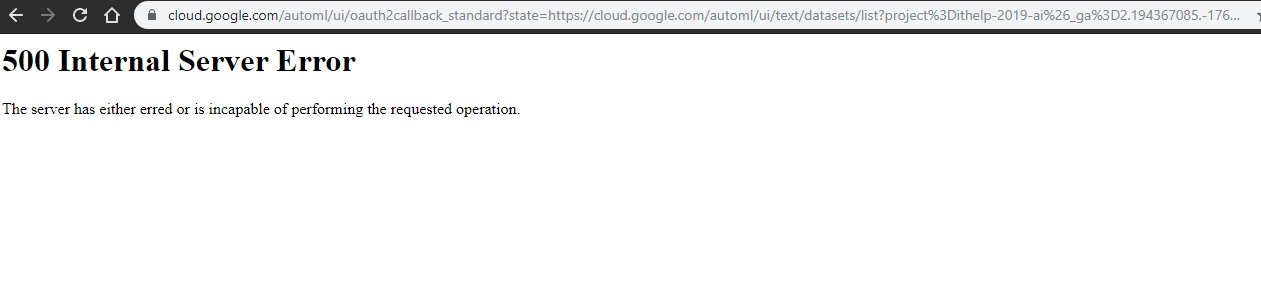
這邊很有意思的是在AutoML Text Classification跟AutoML Sentiment Analysis(圖中兩個紅色的項目)在寫文章的時候,如果你驗證的不是google的第一個帳號,而是選其他帳號登入的話,允許授權後會一直跳500錯誤。
沒關係,這部份我們就選AutoML Entity Extraction,就玩他這個功能,至少他的授權是成功的。
然後我們就照這份Quickstarts來操作,裡面也有範例檔案gs://cloud-ml-data/NL-entity/dataset.csv
那我們就來New一個dataset吧!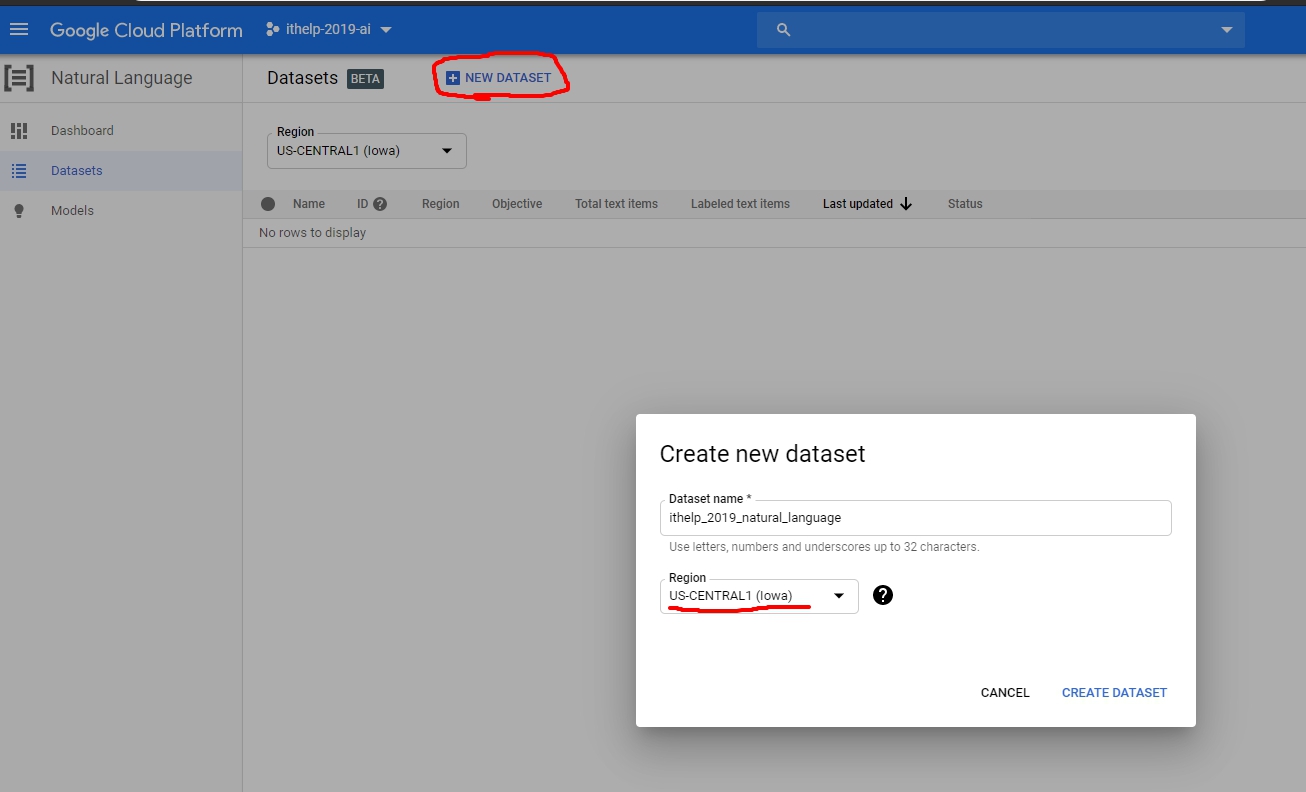
Import是吃一個CSV檔案,也有提供檔案格式,我們載下來的檔案長這個樣子
1 | TRAIN,gs://cloud-ml-data/NL-entity/train.jsonl |
前面的SET可加可不加,但後面要個別帶入jsonl的資料集。輸入後長下面這樣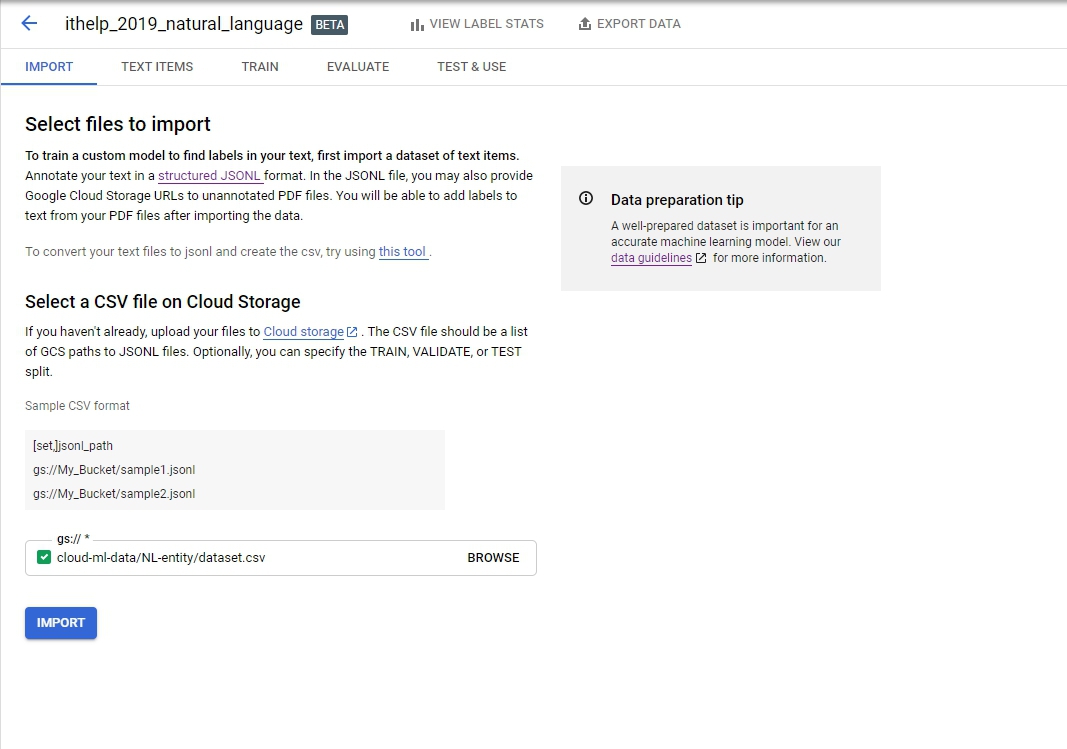
經過一段時間的Processing text items(大概10分鐘),就會有結果
至於這些label怎麼來的?我們就來看看jsonl裡面的內容吧。
1 | {"annotations": [{"text_extraction": {"text_segment": {"end_offset": 54, "start_offset": 27}}, "display_name": "SpecificDisease"}, {"text_extraction": {"text_segment": {"end_offset": 173, "start_offset": 156}}, "display_name": "SpecificDisease"}, {"text_extraction": {"text_segment": {"end_offset": 179, "start_offset": 176}}, "display_name": "SpecificDisease"}, {"text_extraction": {"text_segment": {"end_offset": 246, "start_offset": 243}}, "display_name": "Modifier"}, {"text_extraction": {"text_segment": {"end_offset": 340, "start_offset": 337}}, "display_name": "Modifier"}, {"text_extraction": {"text_segment": {"end_offset": 698, "start_offset": 695}}, "display_name": "Modifier"}], "text_snippet": {"content": "1301937\tMolecular basis of hexosaminidase A deficiency and pseudodeficiency in the Berks County Pennsylvania Dutch.\tFollowing the birth of two infants with Tay-Sachs disease ( TSD ) , a non-Jewish , Pennsylvania Dutch kindred was screened for TSD carriers using the biochemical assay . A high frequency of individuals who appeared to be TSD heterozygotes was detected ( Kelly et al . , 1975 ) . Clinical and biochemical evidence suggested that the increased carrier frequency was due to at least two altered alleles for the hexosaminidase A alpha-subunit . We now report two mutant alleles in this Pennsylvania Dutch kindred , and one polymorphism . One allele , reported originally in a French TSD patient ( Akli et al . , 1991 ) , is a GT-- > AT transition at the donor splice-site of intron 9 . The second , a C-- > T transition at nucleotide 739 ( Arg247Trp ) , has been shown by Triggs-Raine et al . ( 1992 ) to be a clinically benign \" pseudodeficient \" allele associated with reduced enzyme activity against artificial substrate . Finally , a polymorphism [ G-- > A ( 759 ) ] , which leaves valine at codon 253 unchanged , is described .\n "}} |
jsonl就是把json一列一列表達的一種格式
這是train.jsonl裡的第一筆資料,就是對content裡的文字把開始跟結束index的範圍框起來,給他一個label。舉例來說
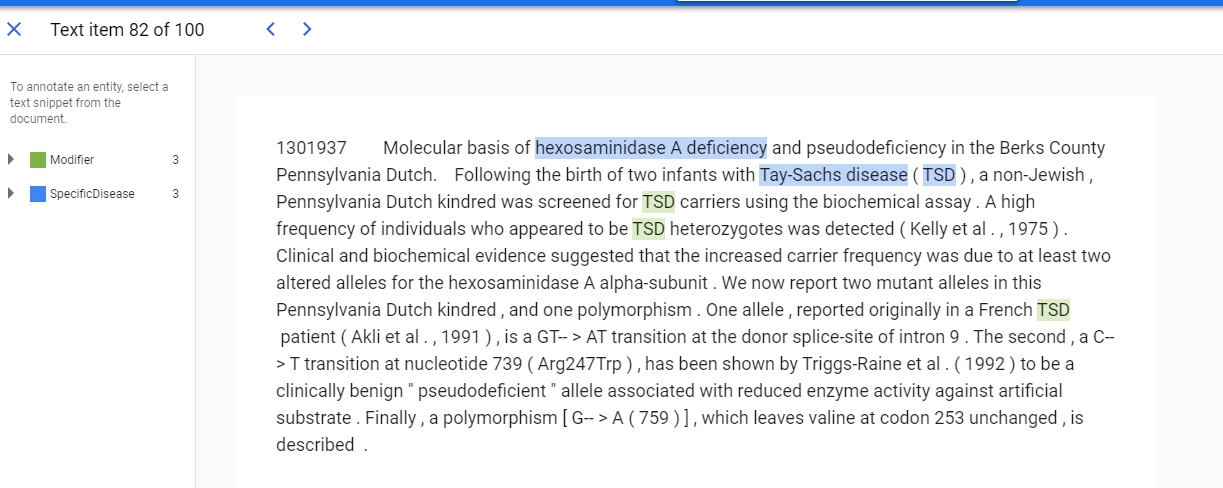
{"end_offset": 54, "start_offset": 27}給他SpecificDisease這個label,而27~54就是hexosaminidase A deficiency這個字,看明白點就是下圖的結果。
好,接下來就train了,訓練前要注意他有建議至少要100個annotation: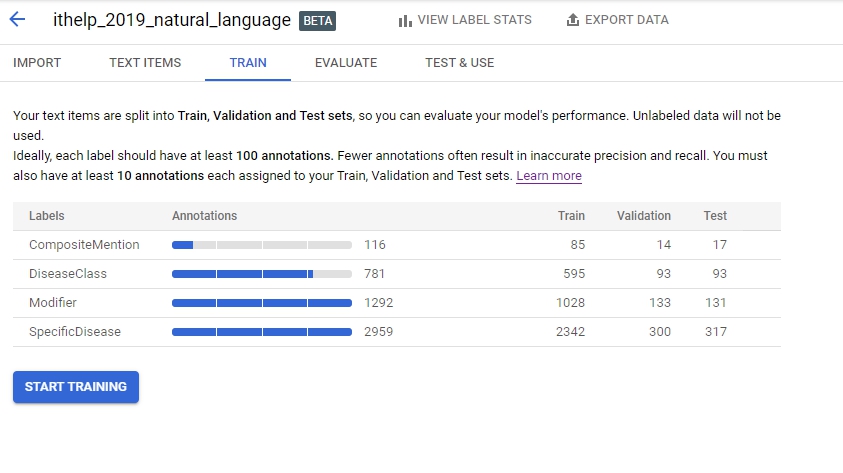
然後又要經過一段時間的訓練…。大概跑了三個小時之久,你才會看到完成的結果。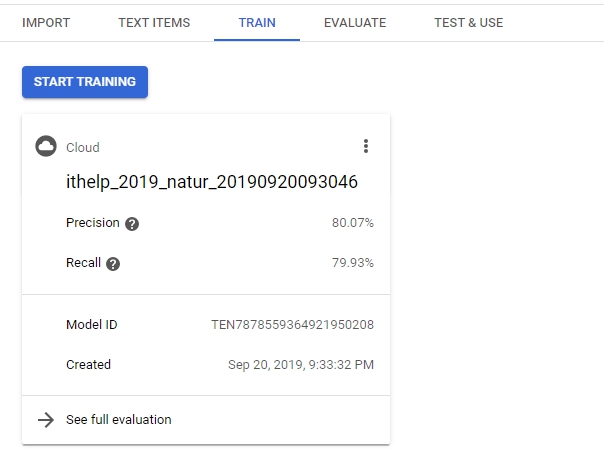
Evaluate的部分可以看到model result,裡面包含recall、precision,還可以看到每個label的true positive、False positives、False negative等等,就可以依照結果去做調整。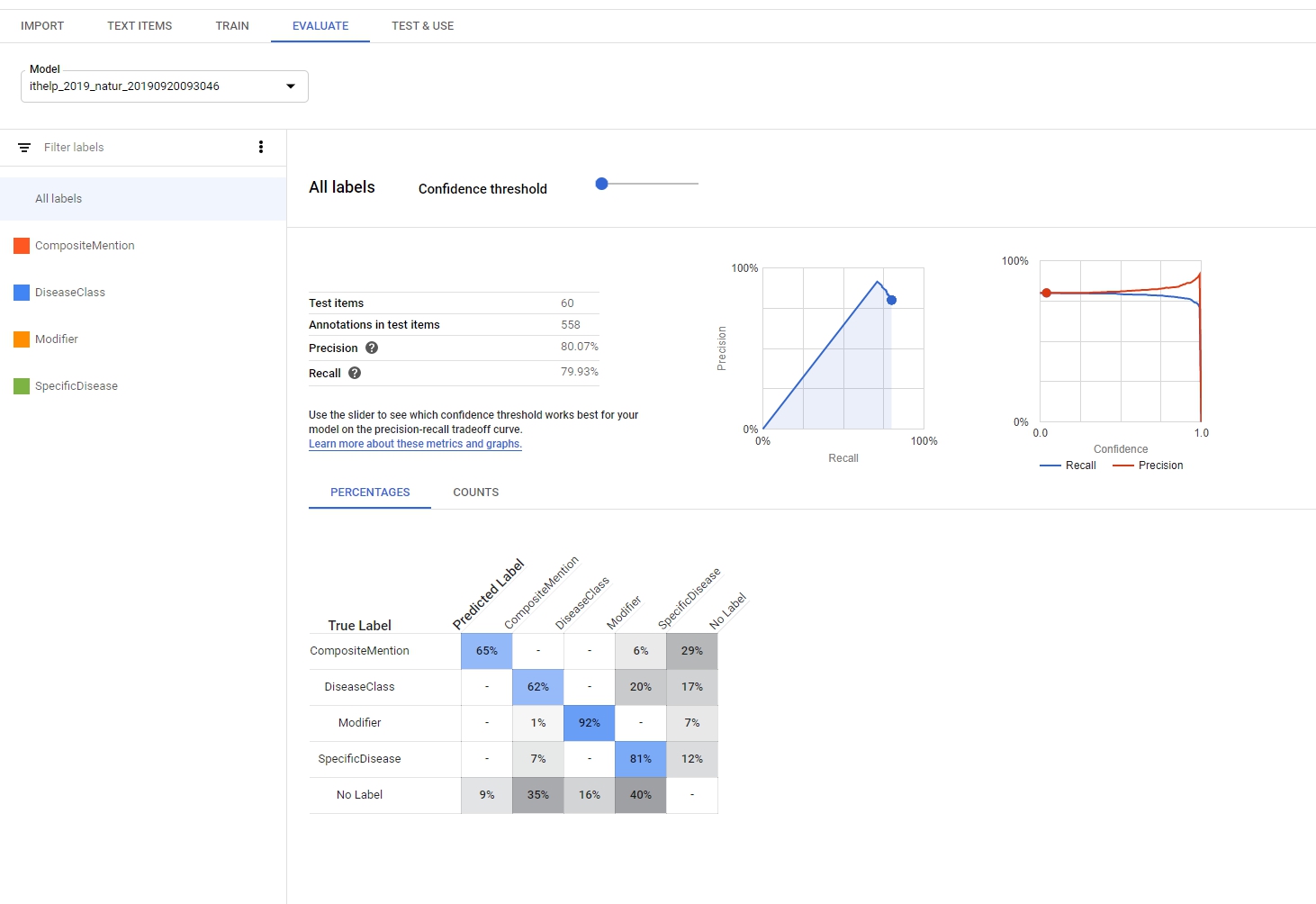
最後到了Test & Use階段,這邊記得要把Model deploy才能做接下來的操作。按下Predict以後就可以看到預測結果了~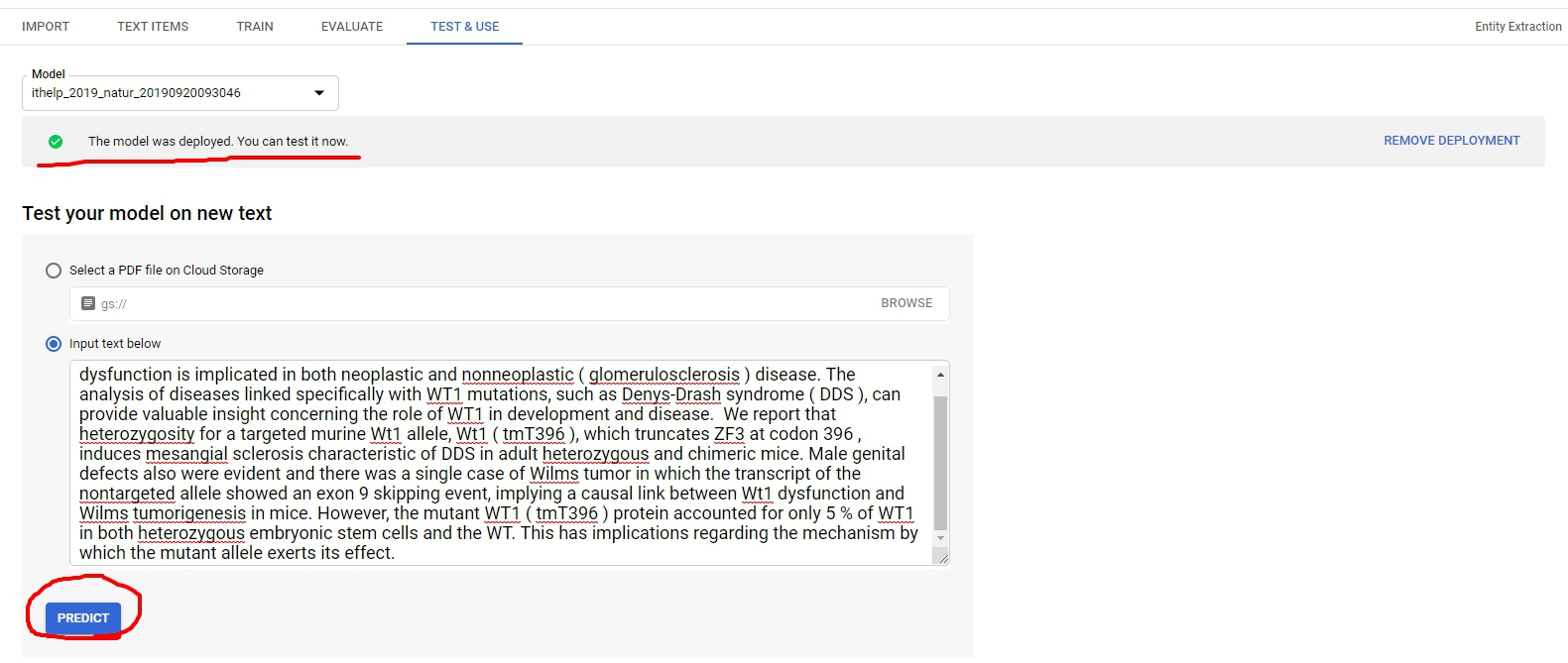

好囉,今天就寫到這邊。model跑得比我想像中的還久,而且又遇到有些GCP服務不能用,很令人尷尬的一次經驗。
OK,謝謝大家的觀看。